Corona virus scare has us all in its grips and as scientists struggle to find a cure for the near pandemic that has us in its jaws, NCBI came to the rescue by handing over the sequence of SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) and creating a separate BLAST database.
So, what is BLAST? BLAST is an acronym for Basic Local Alignment Search Tool introduced by National Centre of Biotechnology Information (NCBI) in 1990, that takes in protein or nucleotide “query’’ (sequence is referred to as query) and compares it to reference sequence in the database and finds regions of similarity and displays results as “Hits’’ with statistical significance. Scientists can compare a new biological sequence (or query) such as an amino-acid sequence of a protein, or DNA / RNA sequences with the library of sequences stored in the BLAST database, and identify the library sequences that match with the query sequence above a certain threshold. The blast software is easy to use and returns the results quickly and accurately.

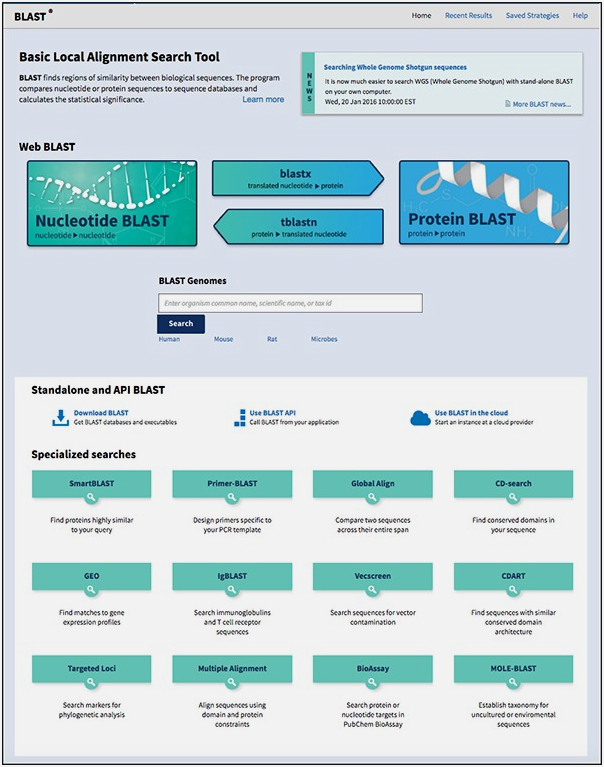
How to access BLAST from NCBI/BLAST website
(Source: https://guides.lib.berkeley.edu/ncbi/blast)
For optimal use of the tool, BLAST offers a myriad of programs. The main types of BLASTs offered by the NCBI tool are:
1) BLASTN: It compares the entered nucleotide sequence against a nucleotide reference sequence in the database.
2) BLASTP: Compares a protein query against a reference protein sequence in the database.
3) BLASTX: Makes comparison of the nucleotide query in 6 translation frames to an amino acid.
4) TBLASTN: This back translates the protein query into all translation frames and then compares to nucleotide sequences in the database.
Then there are several other programs including MEGA BLAST that compares 2 or more nucleotide sequence in database in closely related species; PSI (Position-Specific Iterate) BLAST and RPS(Reverse Position Specific) BLAST are complementary to each other and BLAST an amino acid query and create a position-specific scoring matrix (PSSM) for the query and comparing it to the PSSM score of other sequences and returning the result. The slight difference between the two is that the RPS blast can be used to find conserved domains in proteins. BLAST can also be employed for primer designing, identifying immunoglobulin structures and sequences, etc.
BLAST homepage with the various tools listed
(Source: https://blast.ncbi.nlm.nih.gov/Blast.cgi)
BLAST has a wide range of applications:
– Comparing your own experimental sequence to the sequence of other species and finding homology
– Identifying possible functions of your protein
– Finding which other genes produce similar protein to the one you have found
– Finding protein domains and conserved domains across protein families and species
– As well as for locating specific sequences for designing drugs and targeted vaccines
There are several tutorials available on the NCBI BLAST website which assist in getting the maximum output from the program.
As we said earlier, the BLAST database has the sequence of SARS-Cov2 coronavirus which is easily accessible by all scientists worldwide. The question is how can the sequence of SARS-CoV-2 help in combating the situation?
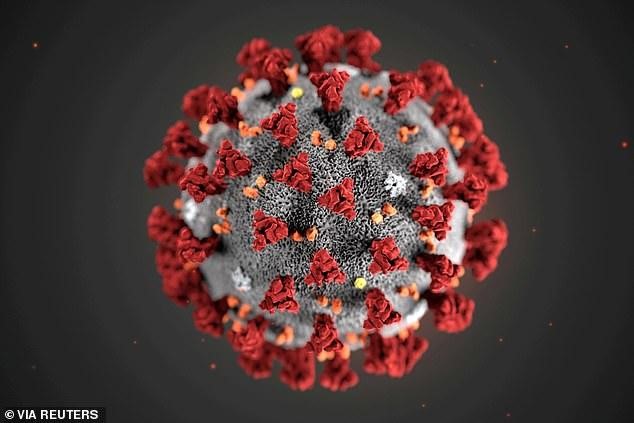
Coronavirus Structure
(Source: https://www.dailymail.co.uk/health/article-7995229/SARS-2-0-Scientists-coronavirus-SARS-CoV-2.html)
Knowing the genomic sequence of the virus will help understand its replication machinery and various parts of its replication cycle. Armed with this knowledge scientists can make antiviral compounds targeting the viral life cycle.
References:
- https://blast.ncbi.nlm.nih.gov/Blast.cgi
- https://bitesizebio.com/21223/how-does-blast-work/
- https://www.slideshare.net/sikhsardari/blast-bioinformatics
- https://www.britannica.com/science/antiviral-drug
- https://www.ncbi.nlm.nih.gov/books/NBK47365/
- https://www.nature.com/articles/d41587-020-00003-1
- https://www.genengnews.com/news/coronavirus-treatment-could-lie-in-existing-drugs/
- https://www.dailymail.co.uk/health/article-7995229/SARS-2-0-Scientists-coronavirus-SARS-CoV-2.html