As nations around the world keep on confronting the inescapable disruption of the COVID-19 pandemic, researchers are hustling to identify diagnostics, immunizations, and medicines. One of the energizing innovations that researches have approached in this crucial scientific endeavour is the gene-altering CRISPR-Cas system. There is no doubt regarding the capability of CRISPR and how it is now having a huge impact on medication, food, biomaterials—essentially our regular day to day lives. With scientists and researchers doing their absolute best to battle the ongoing novel COVID episode they have found CRISPR-based tests for viral discovery.
What is CRISPR?
CRISPR is the acronym for Clustered Regularly Interspaced Short Palindromic Repeats. The CRISPR associated Cas System is the defense mechanism in Bacteria and Archaea that works in the form of a prokaryotic adaptive immune system. CRISPR-Cas frameworks are spread broadly in Bacteria and Archaea providing a shield against viral diseases. It is made of 2 genetic bodies i.e. the CRISPR loci including spacers and repeats and operons of Cas genes. At the point when a virus contaminates a bacterium, it fuses short viral DNA pieces into its chromosome at the CRISPR locus. To counter the virus attack the infected bacterium first transcribes into pre-CRISPR RNA. The pre-CRISPR RNA is then created crRNA i.e. the CRISPR RNA with the help of ribonucleases. The crRNA and the Cas protein shape a network that focuses on identifying the target sequence and generates double-strand breaks in the viral genome. Being complementary to the crRNA, the Cas protein guided by the crRNA recognizes the target sequence in the phage DNA after which the Cas endonuclease cleaves off the viral DNA thus providing protection from viral attacks.
The CRISPR-Cas framework has ended up being a cumulative engineering tool in recent years for bringing about exact and controlled genetic adjustments. Presently there are 2 classes of CRISPR-Cas systems: Class 1 and Class 2. Each of these classes is further subdivided into different types i.e. Class 1 consists of Type I, Type III, and Type IV, and Class 2 is divided into Type II, Type V and Type VI.
How does CRISPR-CAS9 system help in the detection of Covid-19?
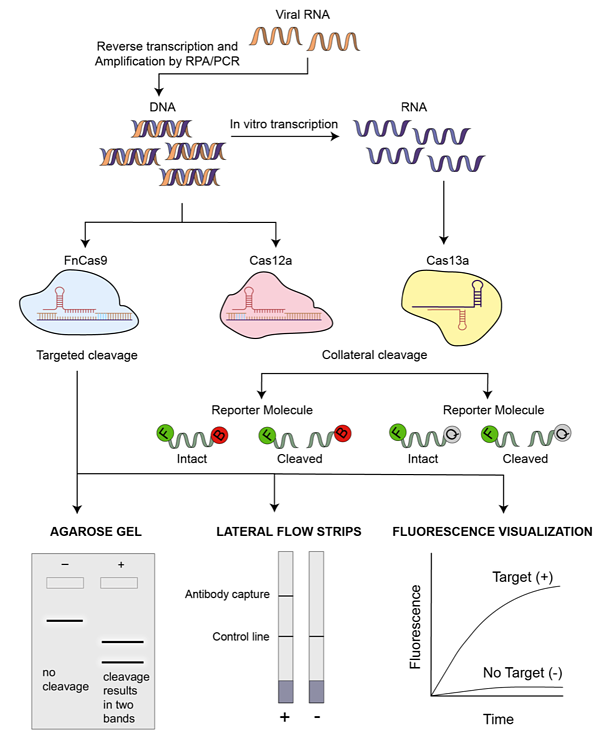
CRISPR picked up prevalence as a genome altering tool after its emergence, but soon the researchers understood its potential in diagnostics. The CRISPR system depends on the guide RNA discovering its complementary target sequence and the regularly utilized Cas 9 nuclease cutting at the exact site. Curiously, on account of the examination on CRISPR’s usefulness as a location-finder, as opposed to the site-explicit cleavage tool, has risen in the previous years. Two nucleases, Cas12a and Cas13a, have been especially famous in CRISPR diagnostics. Cas12a is DNA-explicit and Cas13a is known to work with RNA. Some portion of the mechanism is like that of Cas9—a guide RNA that is complementary to the target sequence is needed for specific binding and the Cas12a/Cas13a nuclease cleaves at the location. An intriguing element of Cas12 and Cas13 nucleases is that they show trans or collateral cutting activity. The nucleases, on finding the target, will also cut other nucleic acid molecules in the region that are non-targeted. This quality is utilized to fabricate reporter frameworks for a visual readout in CRISPR diagnostics.
SHERLOCK: CRISPR-CAS13 Dependent Diagnostic Test
SHERLOCK stands for Specific High-sensitivity Enzymatic Reporter unLOCKing. The SHERLOCK test involves a simple paper strip to indicate the test results. The paper strip is required to be immersed in a processed sample thereafter producing a line indicating the presence or absence of the target molecule. SHERLOCK gives a low budget, simple-to-utilize, and sensitive analytic strategy for identifying the nucleic acid material —a virus, tumor DNA, and numerous different targets. At the center of SHERLOCK’s prosperity is a CRISPR-related protein called Cas13, which can be modified to bind to a particular piece of RNA. Since Cas13a works with RNA it is much convenient in detecting the SARS-CoV-2. Cas13’s objective can be any genetic arrangement that includes viral genomes, changes that cause malignant growth, or genes that present anti-microbial resistance in bacteria. SHERLOCK’s capabilities depend on extra synthetic RNA strands that are utilized to make a signal in the wake of being cleaved. Cas13 will slash up this RNA after it hits its unique original target, delivering the signaling particle, which brings about a readout that shows the presence or absence of the target.
DETECTR Platform for the Detection of Coronavirus
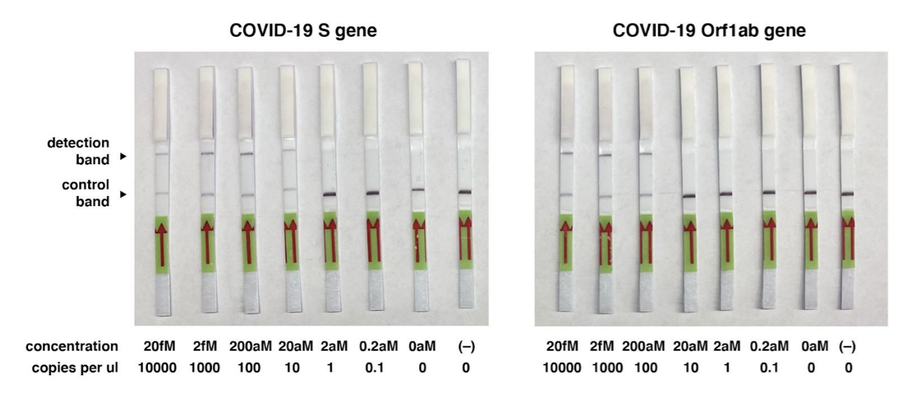
Another method for coronavirus detection is the DNA endonuclease-targeted CRISPR trans reporter (DETECTR), for sensitive detection of DNA by utilizing the Cas12a gene. DETECTR test could be adjusted to distinguish SARS-CoV-2 RNA in around 30 minutes. This platform consists of a combination of Cas12a, its guide RNA, a fluorescent reporter molecule, and recombinase polymerase amplification in order to produce a single reaction. RPA quickly multiples the number of duplicates of the target DNA when warmed to body temperature, increasing the chances of Cas12a to recognize one of them and bind to them thereafter releasing single-strand DNA cutting, bringing about a fluorescent readout. Such a quick diagnostic approach would be especially significant in high-risk regions, for example, air terminals and clinics. The affectability scope of the DETECTR stage is 70-300 duplicates/µl input. It involves 3 steps:
- RNA extricated from test utilizing reverse transcription loop-mediated isothermal amplification (RT-LAMP) is amplified.
- DNA is detected by utilizing Cas12a and crRNA targeting on particular sequences.
- Visual readout utilizing commercially accessible paper strips.
The Return of ‘FELUDA’ to Detect the Novel Coronavirus
FNCAS9 Editor-Limited Uniform Detection Assay- “FELUDA”, produced by Debojyoti Chakraborty and Souvik Maiti at the Council of Scientific and Industrial Research’s constituent lab, the Institute of Genomics and Integrative Biology (CSIR-IGIB) in New Delhi is an indigenous COVID-19 detecting mechanism that utilizes the CRISPR-Cas system which is well known for its cutting-edge technology utilized for the discovery of gene editing and genomic sequences. As per the Ministry of Science and Technology, it adjusts the Cas9 protein to distinguish the Coronavirus during the test. FELUDA test is the world’s first demonstrative test to send a uniquely adjusted Cas9 protein to effectively recognize the virus that results in the COVID-19. The test accomplishes precision levels of conventional RT-PCR tests with faster turnaround time, more affordable equipment, and better convenience. The Tata Group in association with CSIR-IGIB and ICMR has worked intimately to produce this top-notch test that will enable the country to increase COVID-19 testing rapidly and monetarily, with a ‘Made in India’ item that is dependable, safe, budget-friendly, and accessible.
Interestingly, FELUDA is a tribute to the beloved detective character created by Bengali filmmaker-author Satyajit Ray.
The Principle behind Feluda Paper Strip Test:
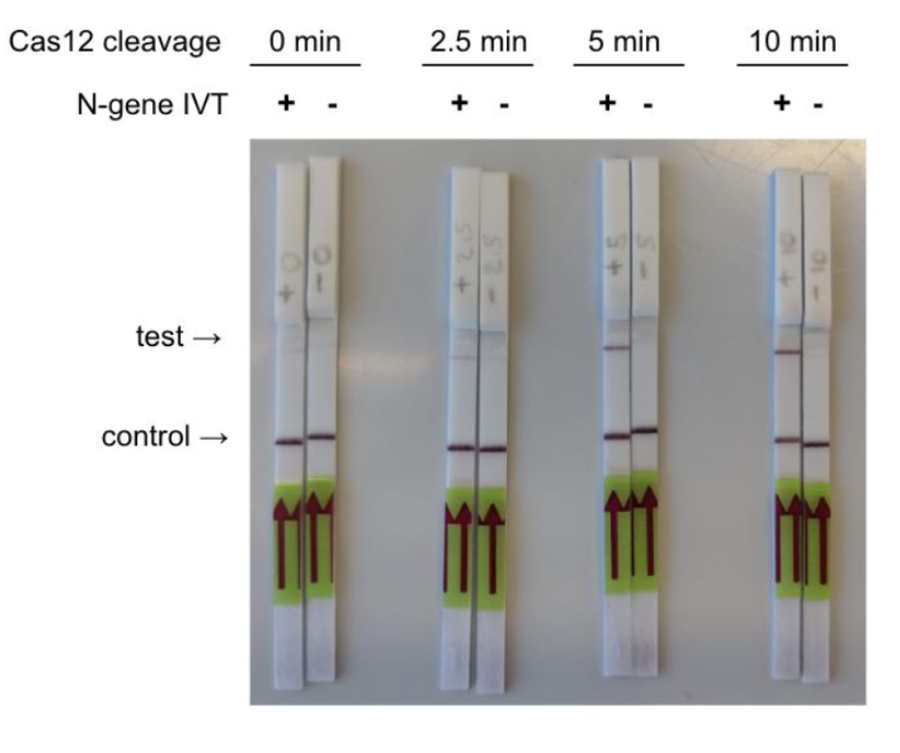
The FELUDA paper strip mechanism utilizes CRISPR-Cas technology to identify and target the genetic material of SARS-CoV2 i.e. the coronavirus. The CRISPR-Cas system recognizes a particular DNA sequence inside the gene, and then, an enzyme that serves the purpose of a molecular scissor is utilized to cut off the DNA sequence, consequently changing the DNA and modifying the gene function. The coronavirus is identified by CRISPR tests that utilize the CAS12 and CAS13 proteins for diagnosis which is why Feluda is the world’s first diagnostic test to utilize a uniquely adjusted Cas9 protein for the effective recognition of the infection. The Cas9 protein is coded in such a way that it causes it to engage with the SARS-CoV2 sequence found in the affected patient’s genetic material. At that point, this Cas9-SARS-CoV2 complex is put on the paper strip with two lines—one control, one test—which helps verify or refute the viral infection.
Working of Feluda Test:
The testing cycle is roughly done in 4 stages:
- Viral RNA is extracted from the nasal and throat swab tests gathered from a suspected patient.
- The viral RNA is changed over into DNA utilizing the RT-PCR (reverse transcriptase-polymerase chain reaction) which additionally does the amplification of the DNA several times, making a few duplicates.
- The Feluda mix containing the Cas9 protein is made, which has the capability to bind to the DNA of the coronavirus. During the amplification, process Chemicals are added to the CRISPR complex and to the DNA to produce a colorimetric response on the paper strip.
- The paper strip is dipped in the Feluda blend which then produces the test result i.e. two lines for a positive and one line on account of a negative test.

The time taken for the determination of the result will be under 30 minutes. As per reports, the expense of the test will approximately be Rs. 500, which will make it considerably more reasonable than the tests that are being utilized right now. The Tata CRISPR test: FELUDA, promoted by CSIR-IGIB (Institute of Genomics and Integrative Biology), has met excellent benchmarks, with 96 percent affectability and 98 percent explicitness for detecting the novel coronavirus.
References:
- https://blog.biomall.in/using-crispr-cas9-in-genome-editing-the-need-of-a-prudent-path/
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7368118/https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7198415/
- https://theprint.in/science/satyajit-rays-feluda-will-soon-detect-coronavirus-in-minutes-thanks-to-csir-scientists/404057/
- https://www.livemint.com/companies/news/tata-group-to-launch-india-s-first-low-cost-covid-19-test-feluda-11600560369240.html
- https://www.aacc.org/publications/cln/cln-stat/2018/march/15/sherlock-detectr-camera-three-new-crispr-technologies
- https://thesocialdigital.com/feluda-covid-19-test-details-and-cost-indias-first-paper-strip-covid-test
- https://zeenews.india.com/india/feluda-first-crispr-covid-19-test-what-is-it-and-how-it-works-explained-2311351.html
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5901762/
- https://www.broadinstitute.org/news/sherlock-team-advances-its-crispr-based-diagnostic-tool