Introduction
The polymerase chain reaction (PCR) is a rapid procedure of in-vitro amplification of DNA that changed the course of molecular biology. This is a revolutionary method developed by Kary B Mullis in 1983, that can generate an ample supply of a specific segment of DNA (an amplicon) from a small amount of starting material (DNA template or target sequence) in few hours. It is based on the principles of complementary nucleic acid hybridization and nucleic acid replication that are applied repeatedly through numerous cycles to result in exponential production of the target sequence
Principle
PCR is primer mediated enzymatic amplification of DNA. It is the DNA polymerase that drives a PCR. A polymerase synthesizes a new strand of DNA complementary to the template strand. The Primer (small pieces of single-stranded DNA) is needed to prime the target DNA sample ready for the DNA polymerase to bind and elongate its 3’ end by adding more nucleotides to generate an extended region of double-stranded DNA.
Steps
The changes in temperature during a cyclic polymerase reaction are used to control the activity of the polymerase and the binding of primers.
Denaturation: In order to begin the reaction, the temperature is raised to 95oC for 15-30 sec. At this temperature, all double-stranded DNA is “denatured” into single strands.
Annealing: The temperature is then rapidly lowered to ~50oC for 20-40 sec. This allows the primers to bind to their complementary sequence in the target DNA.
Extension: The optimal temperature for the polymerase to operate is 72oC. So, in this step polymerase enzyme sequentially adds bases to 3’ end of each primer to extend the DNA sequence.
With one cycle of changing temperatures (95oC, 50oC and 72oC) single DNA segment (single-stranded) amplified into two segments of double-stranded DNA. These two segments are then available for the next amplification cycle. At least 25 to 30 cycles are required to achieve required levels of target sequences in mammalian DNA templates.
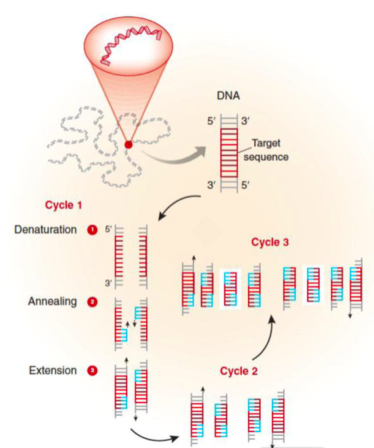
(Garibyan, L., & Avashia, N. (2013). Polymerase chain reaction. The Journal of investigative dermatology, 133(3), 1-4)
Components of PCR
- Thermostable DNA polymerase: The DNA polymerase is required to catalyze template-dependent DNA synthesis. Depending on the efficiency and fidelity to synthesize large DNA products, a wide choice of polymerases is available. For routine laboratory PCRs, Taq polymerase remains the first choice. It polymerizes at the rate of 150 nucleotides per sec per enzyme molecule but lacks 3′ to 5′ exonuclease activity.
- Primers (synthetic oligonucleotides):
- Deoxynucleotide triphosphates(dNTPs): For standard PCR equimolar concentration of all four dNTPs are required. Each dNTP in concentrations of 200-250μM is recommended for Taq polymerase in reactions containing 1.5mM MgCl2
- Divalent Cations: The DNA polymerases require free divalent cations- usually Mg2+ for its activity. As dNTPs and oligonucleotides also bind to Mg2+, therefore, the molar concentration of the cation must exceed the molar concentration of phosphate groups contributed by dNTPs and primers. Routinely, 1.5 mM of Mg2+ concentration is used. In order to decrease nonspecific priming the concentration of Mg2+ is recommended to increase 4.5 mM or 6mM. Therefore, the optimal concentration of Mg2+ should be empirically determined for each combination of primers and template
- Buffer: Tris-Cl, adjusted to a pH between 8.3 and 8.8 at room temperature is included in standard PCRs at a concentration of 10mM in order to maintain pH of the reaction mixture.
- Monovalent cations: For standard PCR buffer, 50mM KCl works well for amplification of segments of DNA >500bp in length
- Template DNA: Template DNA containing target sequences can be added to PCR in single or double-stranded form.
Analysis of the PCR product
The PCR product can be visualized using two methods:
(1) Agarose gel electrophoresis, which separates DNA products on the basis of size and charge. It is the easiest method to visualize PCR products and determine the presence and the size of the product.
(2) Labelling the PCR primers or nucleotides with fluorescent dyes prior to PCR amplification. This allows the labels to be directly incorporated into the PCR product.
Visualization of PCR product using Agarose gel electrophoresis
(Source: Garibyan, L., & Avashia, N. (2013). Polymerase chain reaction. The Journal of investigative dermatology, 133(3), 1-4)
Limitations of PCR
- The DNA polymerase lacks 3’ to 5’ exonuclease activity thus prone to errors and can lead to mutations in the generated fragment.
- The nonspecific binding of the primers to other similar sequences on the template DNA may alter the specificity of the generated PCR product
- Some prior sequence information of the target sequence is necessary to design primers to generate a PCR product
References:
- Mullis KB. The unusual origin of the polymerase chain reaction. Scientific American. 1990;262(4):56–61. 64–5
- Garibyan, L., & Avashia, N. (2013). Polymerase chain reaction. The Journal of investigative dermatology, 133(3), 1-4
- Staněk L. [Polymerase chain reaction: basic principles and applications in molecular pathology]. Cesk Patol. 2013 Jun;49(3):119-21